RMI
This is a reference implementation of recursive model indexes (RMIs). A prototype RMI was initially described in The Case for Learned Index Structures by Kraska et al. in 2017.
RMI basics
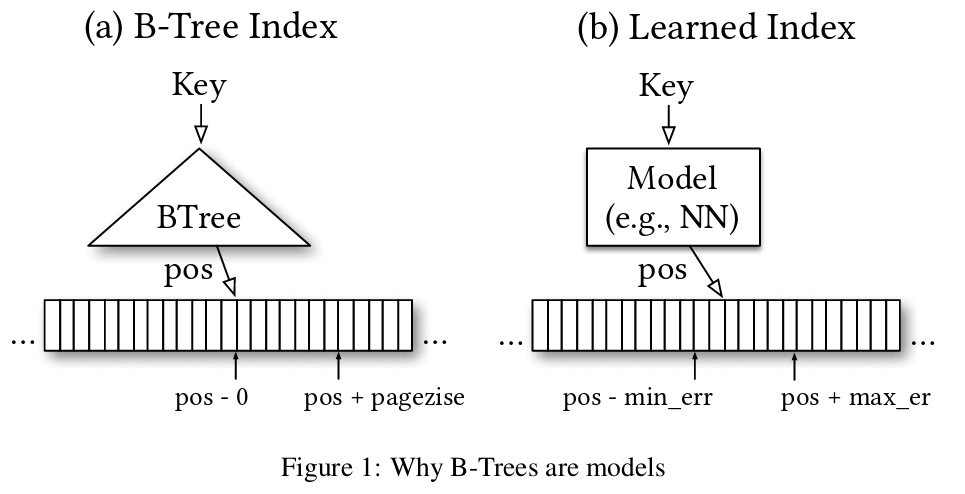
Like binary search trees, an RMI is a structure to help search through sorted data. Given a sorted array, an RMI is a function that maps a key to an approximate index. This approximate index can be used as a starting point for a linear, exponential, or binary search. The SOSD benchmark demonstrates that RMIs can outperform binary search and many other standard approaches as well.
Unlike a binary search tree, an RMI uses machine learning techniques to build this approximation function. The result is normally a small, compact mathematical function that can be evaluated quickly. RMIs are a good tool when you need to search the same sorted data many times. Compared to other structures, RMIs:
- (
➕ ) Offer faster lookup times (when properly tuned) - (
➕ ) Are generally much smaller than traditional structures like B-Trees or radix trees - (
➖ ) Must be trained ahead of time on a dataset - (
➖ ) Do not support inserts (without retraining the model)
Many more details can be found in the original paper.
Using this implementation
To use the reference implementation, clone this repository and install Rust.
The reference RMI implementation is a compiler. It takes a dataset as input, and produces C/C++ source files as outputs. The data input file must be a binary file containing:
- The number of items, as a 64-bit unsigned integer (little endian)
- The data items, either 32-bit or 64-bit unsigned integers (little endian)
If the input file contains 32-bit integers, the filename must end with uint32. If the input file contains 64-bit integers, the filename must end with uint64. If the input file contains 64-bit floats, the filename must end with f64.
In addition to the input dataset, you must also provide a model structure. For example, to build a 2-layer RMI on the data file books_200M_uint32 (available from the Harvard Dataverse) with a branching factor of 100, we could run:
cargo run --release -- books_200M_uint32 my_first_rmi linear,linear 100
Logging useful diagnostic information can be enabled by setting the RUST_LOG environmental variable to trace: export RUST_LOG=trace.
Generated code
The RMI generator produces C/C++ source files in the current directory. The command directly above, for example, produces the following output. The C/C++ sources contain a few publicly-exposed fields:
#include <cstddef>
#include <cstdint>
namespace wiki {
bool load(char const* dataPath);
void cleanup();
const size_t RMI_SIZE = 50331680;
const uint64_t BUILD_TIME_NS = 14288421237;
const char NAME[] = "wiki";
uint64_t lookup(uint64_t key, size_t* err);
}
- The
RMI_SIZEconstant represents the size of the constructed model in bytes. - The
BUILD_TIME_NSfield records how long it took to build the RMI, in nanoseconds. - The
NAMEfield is a constant you specify (and always matches the namespace name). - The
loadfunction will need to be called before any calls tolookup. ThedataPathparameter must the path to the directory containing the RMI data (rmi_datain this example / the default). - The
lookupfunction takes in an unsigned, 64-bit integer key and produces an estimate of the offset. Theerrparameter will be populated with the maximum error from the RMI's prediction to the target key. This lookup error can be used to perform a bounded binary search. If the error of the trained RMI is low enough, linear search may give better performance.
If you run the compiler with the --no-errors flag, the API will change to no longer report the maximum possible error of each lookup, saving some space.
uint64_t lookup(uint64_t key);
RMI Layers and Tuning
Currently, the following types of RMI layers are supported:
linear, simple linear regressionlinear_spline, connected linear spline segmentscubic, connected cubic spline segmentsloglinear, simple linear regression with a log transformnormal, normal CDF with tuned mean, variance, and scale.lognormal, normal CDF with log transformradix, eliminates common prefixes and returns a fixed number of significant bits based on the branching factorbradix, same as radix, but attempts to choose the number of bits based on balancing the datasethistogram, partitions the data into several even-sized blocks (based on the branching factor)
Tuning an RMI is critical to getting good performance. A good place to start is a cubic layer followed by a large linear layer, for example: cubic,linear 262144. For automatic tuning, try the RMI optimizer using the --optimize flag:
cargo run --release -- --optimize optimizer_out.json books_200M_uint64
By default, the optimizer will use 4 threads. If you have a big machine, consider increasing this with the --threads option.
The optimizer will output a table, with each row representing an RMI configuration. By default, the optimizer selects a small set of possible configurations that are heuristically selected to cover the Pareto front. Each column contains:
Models: the model types used at each level of the RMIBranch: the branching factor of the RMI (number of leaf models)AvgLg2: the average log2 error of the model (which approximates the number of binary search steps required to find a particular key within a range predicted by the RMI)MaxLg2: the maximum log2 error of the model (the maximum number of binary search steps required to find any key within the range predicted by the RMI)Size (b): the in-memory size of the RMI, in bytes.
Citation and license
If you use this RMI implementation in your academic research, please cite the CDFShop paper:
Ryan Marcus, Emily Zhang, and Tim Kraska. 2020. CDFShop: Exploring and Optimizing Learned Index Structures. In Proceedings of the 2020 ACM SIGMOD International Conference on Management of Data (SIGMOD '20). Association for Computing Machinery, New York, NY, USA, 2789–2792. DOI:https://doi.org/10.1145/3318464.3384706
If you are comparing a new index structure to learned approaches, or evaluating a new learned approach, please take a look at our [benchmark for learned index structures](https://learned.systems/sosd.
For RMIs and learned index structures in general, one should cite the original paper:
Tim Kraska, Alex Beutel, Ed H. Chi, Jeffrey Dean, and Neoklis Polyzotis. 2018. The Case for Learned Index Structures. In Proceedings of the 2018 International Conference on Management of Data (SIGMOD '18). Association for Computing Machinery, New York, NY, USA, 489–504. DOI:https://doi.org/10.1145/3183713.3196909
This work is freely available under the terms of the MIT license.