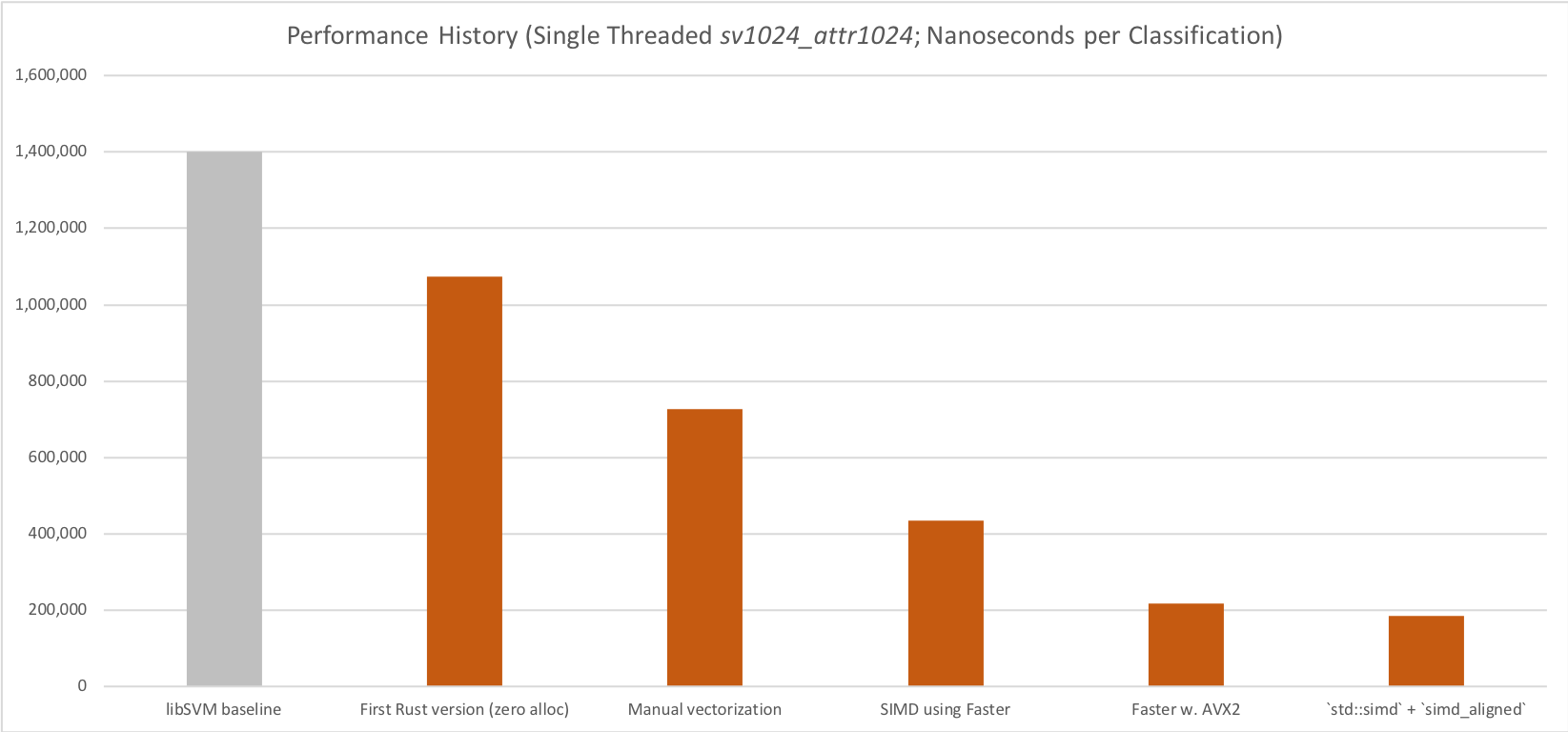In One Sentence
You trained a SVM using libSVM, now you want the highest possible performance during (real-time) classification, like games or VR.
Highlights
- loads almost all libSVM types (C-SVC, ν-SVC, ε-SVR, ν-SVR) and kernels (linear, poly, RBF and sigmoid)
- produces practically same classification results as libSVM
- optimized for SIMD and can be mixed seamlessly with Rayon
- written in 100% Rust
- allocation-free during classification for dense SVMs
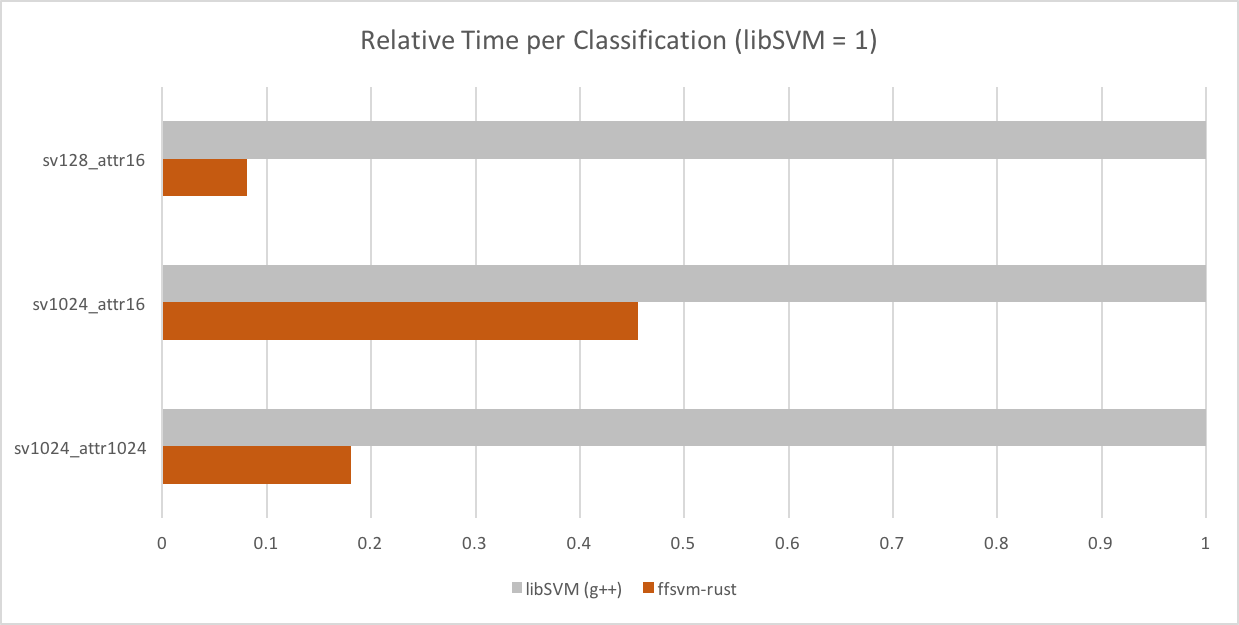
- 2.5x - 14x faster than libSVM for dense SVMs
- extremely low classification times for small models (e.g., 128 SV, 16 dense attributes, linear ~ 500ns)
- successfully used in Unity and VR projects (Windows & Android)
Note: Currently requires Rust nightly (March 2019 and later), because we depend on RFC 2366 (portable SIMD). Once that stabilizes we'll also go stable.
Usage
Train with libSVM (e.g., using the tool svm-train), then classify with ffsvm-rust.
From Rust:
// Replace `SAMPLE_MODEL` with a `&str` to your model.
let svm = DenseSVM::try_from(SAMPLE_MODEL)?;
let mut problem = Problem::from(&svm);
let features = problem.features();
features[0] = 0.55838;
features[1] = -0.157895;
features[2] = 0.581292;
features[3] = -0.221184;
svm.predict_value(&mut problem)?;
assert_eq!(problem.solution(), Solution::Label(42));
Status
- June 7, 2019: Gave up on 'no
unsafe', but gained runtime SIMD selection. - March 10, 2019: As soon as we can move away from nightly we'll go beta.
- Aug 5, 2018: Still in alpha, but finally on crates.io.
- May 27, 2018: We're in alpha. Successfully used internally on Windows, Mac, Android and Linux on various machines and devices. Once SIMD stabilizes and we can cross-compile to WASM we'll move to beta.
- December 16, 2017: We're in pre-alpha. It will probably not even work on your machine.
Performance
Classification time vs. libSVM for dense models.
Performance milestones during development.
All performance numbers reported for the DenseSVM. We also have support for SparseSVMs, which are slower for "mostly dense" models, and faster for "mostly sparse" models (and generally on the performance level of libSVM).
Tips
- For an x-fold performance increase, create a number of
Problemstructures, and process them with Rayon'spar_iter.